Andrey lab: epigenetic control of developmental processes
During embryogenesis, the activation and repression of genes in time and in space instruct the fabric of organs and structures. The molecular processes that orchestrate these activities are thus essential for the development of the organism, and their perturbations can lead to malformations or diseases.
Konrad Guenther, Vom Urtier zum Menschen, 1909.
Our laboratory focuses on understanding the molecular processes from which the gene expression patterns required for the normal embryonic development stem. Currently, our research is concentrated on two synergistic lines of research:
- We aim at understanding how regulatory regions and genes communicate with each other during development. In particular, we focus on how spatial proximity between these elements is established and drives gene transcriptional onset, ultimately forming organs and structures.
- We aim at characterizing the transcriptomic identity and chromatin topologies of the specific cell populations that constitute developing organs in normal and mutant genetic backgrounds.
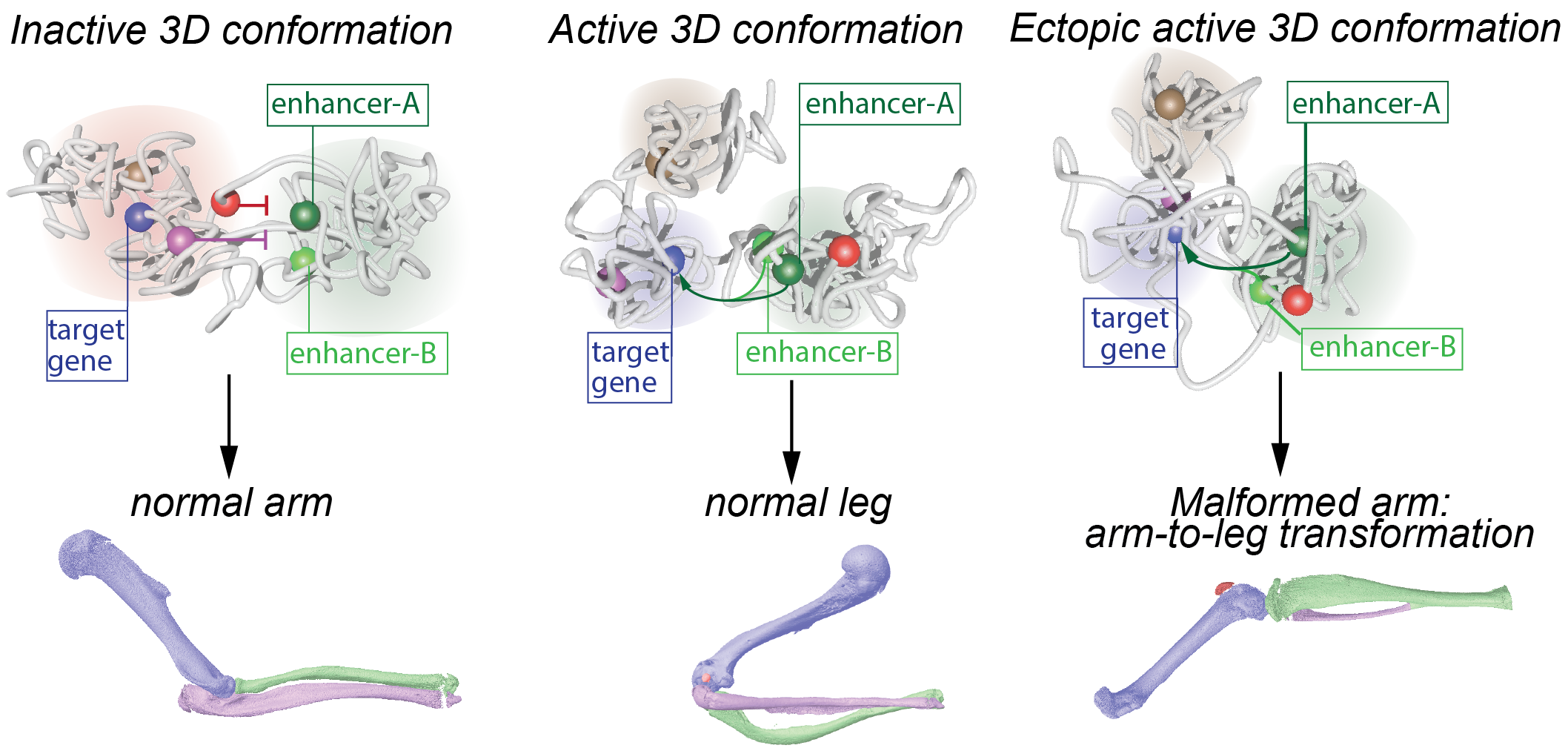
Changes in 3D genome conformation can induce ectopic gene expression and lead to leg malformations (Adapted from Kragesteen et al., Nat Genet., 2018)
To address these aims, our lab, in collaboration with the institute's transgenic facility uses tetraploid complementation, which enables to produce embryos from wildtype and genetically engineered Embryonic Stem Cells (ESCs), while drastically limiting the number of used animals. We characterize these embryos with high-resolution mapping of chromatin interactions and modifications. Ultimately, our objective is to establish the molecular rules that govern the relationship between chromatin biology, gene expression and normal/abnormal embryonic development over developmental time.

Experimental workflow to characterize genome structure, chromatin modifications and cell-type specific transcriptomics in wildtype and genetically-engineered embryos
Selected Publications
Chondrogenic Enhancer Landscape of Limb and Axial Skeleton Development
Darbellay F., Ramisch A., Lopez-Delisle L., Koiscki M., Visel A. and Andrey G.
Preformed Chromatin Topology Assists Transcriptional Robustness of Shh during Limb Development.
Proc Natl Acad Sci U S A. 2019 May 30. pii: 201900672. doi: 10.1073/pnas.1900672116
Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis.
Kragesteen BK, Spielmann M, Paliou C, Heinrich V, Schöpflin R, Esposito A, Annunziatella C, Bianco S, Chiariello AM, Jerković I, Harabula I, Guckelberger P, Pechstein M, Wittler L, Chan WL, Franke M, Lupiáñez DG, Kraft K, Timmermann B, Vingron M, Visel A, Nicodemi M, Mundlos S, Andrey G.
Nat Genet. 2018 Oct;50(10):1463-1473. doi: 10.1038/s41588-018-0221-x. Epub 2018 Sep 27.
Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding.
Andrey G, Schöpflin R, Jerković I, Heinrich V, Ibrahim DM, Paliou C, Hochradel M, Timmermann B, Haas S, Vingron M, Mundlos S.
Genome Res. 2017 Feb;27(2):223-233. doi: 10.1101/gr.213066.116. Epub 2016 Dec 6.
Formation of new chromatin domains determines pathogenicity of genomic duplications.
Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, Spielmann M, Timmermann B, Wittler L, Kurth I, Cambiaso P, Zuffardi O, Houge G, Lambie L, Brancati F, Pombo A, Vingron M, Spitz F, Mundlos S.
Nature. 2016 Oct 13;538(7624):265-269. doi: 10.1038/nature19800. Epub 2016 Oct 5.
Deletions, Inversions, Duplications: Engineering of Structural Variants using CRISPR/Cas in Mice.
Kraft K, Geuer S, Will AJ, Chan WL, Paliou C, Borschiwer M, Harabula I, Wittler L, Franke M, Ibrahim DM, Kragesteen BK, Spielmann M, Mundlos S, Lupiáñez DG, Andrey G.
Cell Rep. 2015 Feb 4. pii: S2211-1247(15)00029-7. doi: 10.1016/j.celrep.2015.01.016.
A switch between topological domains underlies HoxD genes collinearity in mouse limbs.
Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D.
Science. 2013 Jun 7;340(6137):1234167. doi: 10.1126/science.1234167.
